Chromophores/Fluorophores: Spectral properties and characteristics
Bachem offers series of peptide-based enzyme substrates linked to chromophores or fluorophores. The advantage of these chromogenic and fluorogenic substrates is the facile spectrometric detection and analysis of the reaction products. The following guide contains some useful information about the various kinds of substrates.
Chromophores
The characteristic feature of chromophores to absorb light of UV and visible wavelengths from 200 nm to 400 nm and from 400 nm to 800 nm, respectively, can be used to determine their concentration by absorption photometry. The method measures the decrease in light intensity when light passes through a colored solution.
The distance which the light has to pass through a solution is called the path length. With a linearly rising concentration of the chromophore solution, the intensity of the emergent beam of light falls off exponentially.
The absorbance (A) is defined as follows:
A = log I0/I = log 1/ T
I = intensity of transmitted light
I0 = intensity of incident light
T = I/I0 = transmittance
Since the absorbance (also called extinction or optical density (O.D.)) is linear to the concentration and the path length the Lambert-Beer’s law can be applied (the law is limited since it is only valid for highly diluted solutions).
Lambert-Beer’s law A = ε x c x d
A = absorbance (dimensionless)
c = concentration mol/l (M)
d = path length cm
ε = molar decadic absorption coefficient l mol-1 cm-1 (M-1 cm-1)*
*(the units for ε found in the literature are usually M-1 x cm-1)
Knowing the molar absorption coefficient of a chromophore in solution its concentration can be calculated according to the Lambert-Beer’s law:
c = A/εx d
Please note that molar absorption coefficients may depend on the temperature, pH, and the ionic strength of a solution.
| Chromophore | Detection Wavelength* | Molar Absorption Coefficient* | |
|---|---|---|---|
| Dnp[1,2,3,4] (2,4-Dinitrophenyl) | 365 nm 354 nm 400 nm 400 nm | ε365 nm = 17 300 M-1cm-1 ε354 nm = 16 300 M-1cm-1 ε400 nm = 6 100 M-1cm-1 ε400 nm = 6 985 M-1cm-1 | |
| FA[5,6] (3-(2-Furyl)acryloyl) (decrease in absorbance measured) | 322-345 nm | ε305 nm = 24 700 M-1cm-1 Δ324 nm = 2 510 M-1cm-1 ε322 nm = 13 400 M-1cm-1 Δ322 nm = 2 300 M-1cm-1 | |
| βNA[7] (β-Naphthylamide) (requires color reaction) | 540 nm (detected by coupling with tetrazotized diorthoanisidine (Diazo Blue B)) 560 nm (by the use of a modified Bratton-Marshall method) | ||
| β-Naphthyl Ester[8] (requires color reaction) | 520 nm (detected by coupling to the zinc-stabilized diazonium salt, Fast Garnet) | ||
| pNA[9,10] (p-Nitroanilide) | 405 nm 410 nm | ε405nm = 9 450 M-1cm-1 ε410 nm = 8 800 M-1cm-1 | |
| Onp[11] (p-Nitrophenyl ester) | 347 nm | ε347 nm = 5 500 M-1cm-1 | |
| SBzl[12] (Thiobenzyl Ester) (requires color eaction) | 324 nm (formation of 4-thiopyridone with 4,4’-dithiodipyridine) 410 nm (formation of 2-nitro-5-thiobenzoate anion with 5,5´-dithiobis-(2-nitrobenzoic acid)) | ε324 nm = 19 800 M-1cm-1 ε410 nm = 14 000 M-1cm-1 |
Table 1: Chromophores
*the values listed are as reported in the cited literature.
2,4-Dinitrophenyl (Dnp) Substrates
Dnp is normally used as a quencher for various fluorophores as indicated in Table 3. With some substrates (e.g. M-1340), however, enzymatic cleavage is assayed by measuring the absorption of the Dnp-linked peptide cleavage product at 365 nm after organic extraction from the assay buffer.
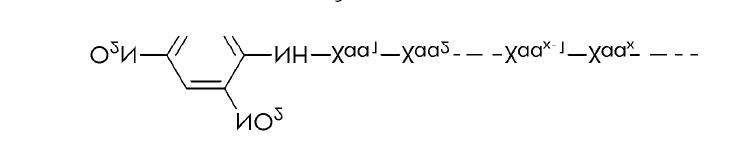
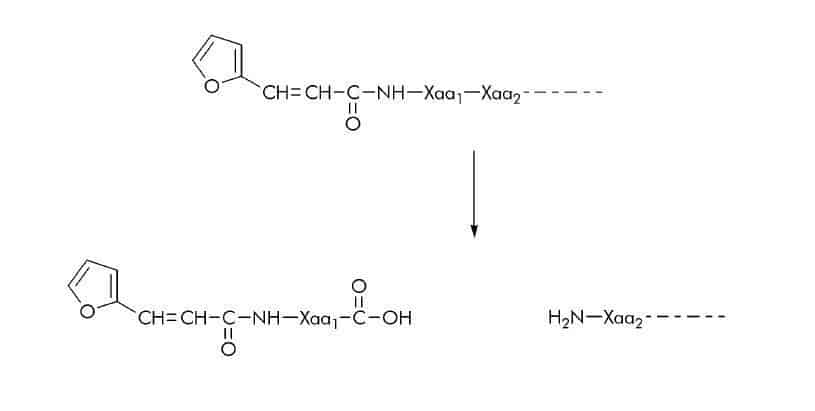
3-(2-Furyl)acryloyl (FA) Substrates
FA-linked substrates have been used for continuous spectrophotometric assays of various proteases such as collagenases, thermolysin, angiotensin I-converting enzyme, elastase, and carboxypeptidase Y. Assays with FA-peptide substrates are based on the blue shift in the near-ultraviolet absorption band of the FA-peptide when the peptide bond between the first and second residue is hydrolyzed. The resulting decrease in absorbance can be measured at wavelengths between 322 nm and 345 nm.
β-Naphthylamide (βNA) Substrates
βNA substrates can either be used as chromogenic or fluorogenic substrates. Photometry, however, requires coupling of the released β-naphthylamine to an azo dye (please see also ‘Fluorophores’).
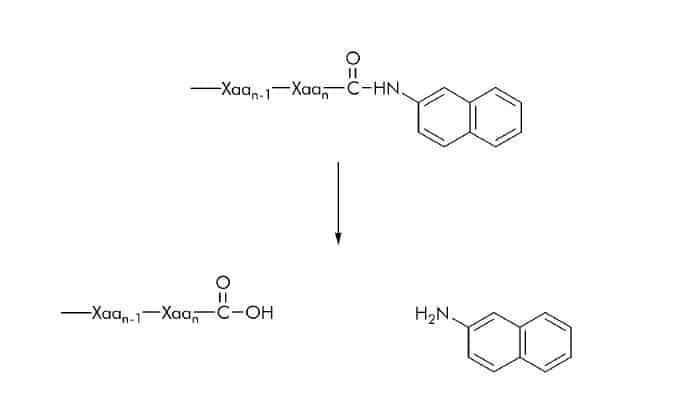
β-Naphthyl Ester Substrates
Hydrolytic activity using β-naphthyl ester substrates can be measured at 520 nm by coupling the liberated naphthol to the zinc-stabilized diazonium salt, Fast Garnet.
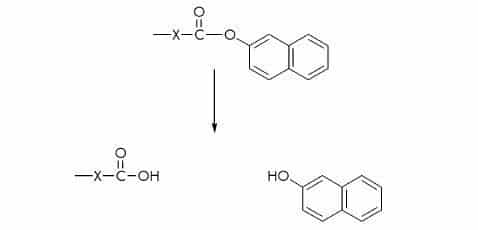
p-Nitroanilide (pNA) Substrates
pNA substrates are commonly used for the measurement of serine and cysteine proteases. The released p-nitroaniline shows a different absorption spectrum than pNA which is linked via an amide bond to the carboxy function of the peptide. p-Nitroaniline is either measured at 405 nm or 410 nm.
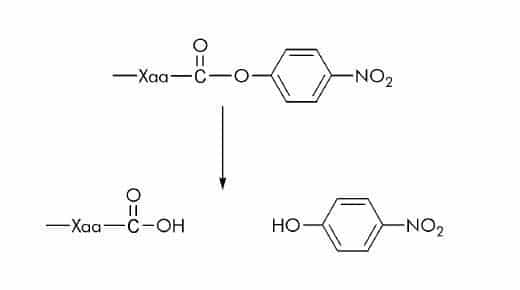
p-Nitrophenyl Ester (ONp) Substrates
In few substrates p-nitrophenyl is linked via an ester bond to the carboxyl group of an amino acid. Esterase activity can be measured spectrophotometrically at 347 nm. At this wavelength the absorption of p-nitrophenol (ε347 nm = 5 500 M-1 cm-1) is independent of the pH.
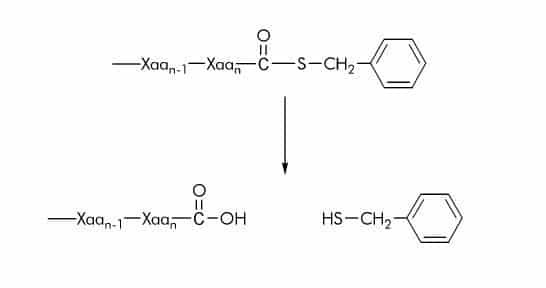
Thiobenzyl Ester (SBzl) Substrates
Many commercially available substrates contain thiobenzylester groups which upon cleavage can be converted into a chromophore by using 5,5´-dithiobis-(2-nitrobenzoic acid) also known as DTNB or Ellman’s reagent. The rate of thioester hydrolysis is measured at 405 nm to monitor the formation of 2-nitro-5-thiobenzoate anion (ε410nm = 14 000 M-1cm-1). Alternatively, 4,4’-dithiodipyridine can be used to measure the thiolation degree by detecting the formation of 4-thiopyridone at 324 nm (ε324nm = 19 800 M-1cm-1).
| Fluorophore Excitation | Excitation Wavelength* | Emission Wavelength* | |
|---|---|---|---|
| Abz [14,15,16] (2-Aminobenzoyl or Anthraniloyl) | 320 nm | 420 nm | |
| N-Me-Abz [17] (N-Methyl-anthraniloyl) | 340 - 360 nm | 440 - 450 nm | |
| AFC[18,19] (7-Amido-4-trifluoromethylcoumarin) | 395 nm 400 nm | 495 nm 505 nm | |
| AMC[20,21,22,10] (7-Amido-4-methylcoumarin) | 360 nm 360 nm 380 nm 380 nm | 440 nm 460 nm 440 nm 460 nm | |
| EDANS [23] (5-[(2-Aminoethyl)-amino]naphthalene-1-sulfonyl) | 340 nm | 490 nm | |
| FITC [24] (Fluorescein isothiocyanate) | 490 nm | 520 nm | |
| 4MβNA [25,26,27,28] (4-Methoxy-β-naphthylamide) | 335 nm 340 nm 340 nm 350 nm | 410 nm 425 nm 430 nm 440 nm | |
| Mca[29] ((7-Methoxycoumarin-4-yl)acetyl) | 325 nm | 392 nm | |
| βNA [30,31] (β-Naphthylamide) | 320 nm 340 nm | 420 nm 410 nm | |
| Trp[14] (Tryptophan) | 280 nm | 360 nm |
Table 2: Fluorophores
*the values listed are as reported in the cited literature.
Fluorescence
Fluorophores are substances which, like chromophores, absorb light in the UV or visible range. In contrast to chromophores they re-emit part of the light as radiation. This process is called fluorescence and can be illustrated by the energy level diagram suggested by A. Jablonski.
Absorption of light (hνA) causes an electron to be promoted from its electronic ground state (designated as S0) to an excited state (usually S1). Every energy state has several vibrational energy levels 0, 1, 2 etc. During the lifetime of the excited state, i.e. the time elapsed between excitation of the molecule and emission of the photon (usually between 1-10 ns) part of the energy is lost by internal vibration.
As a result the wavelength of the emitted light (hνF) is always longer than that of the exciting light. This phenomenon is called the Stokes shift and allows the detection of emission against a background of light derived from excitation. Usually, the fluorescence excitation spectrum of a fluorophore in a diluted solution is identical to its absorption spectrum and under the same conditions, the fluorescence emission spectrum is independent of the excitation wavelength.
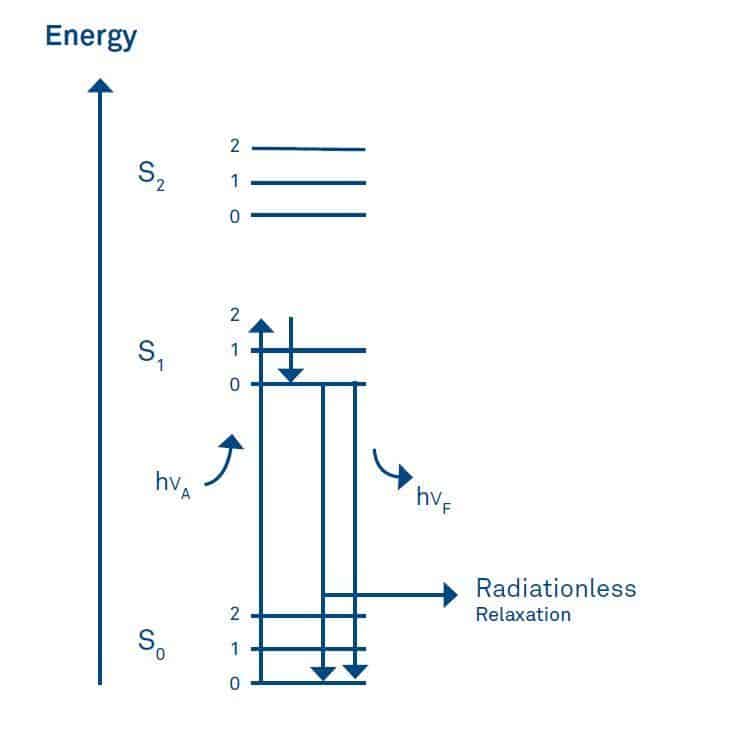
In a diluted solution, fluorescence intensity is linearly proportional to several parameters as deduced from the Lambert-Beer’s law. These are the molar absorption coefficient, the path length, the intensity of the incident light, and the quantum yield which is the ratio of the number of emitted to the total number of absorbed photons.
Fluorescence detection is dependent on the sensitivity of the instrument and is therefore measured in arbitrary units. Higher concentrations of the fluorophore (> 0.1 absorption units) lead to deviations from linearity due to loss of excitation intensity across the cuvette path length as the excitation light is absorbed by the fluorophore. This phenomenon is known as the inner filter effect.
Other effects which influence fluorescence measurements are related to intrinsic or background fluorescence originating from sample preparations and buffer contaminants, respectively. To minimize fluorescence derived from contaminants it is recommended to use materials of the highest purity. Fluorescence spectra may also be dependent on the solvent.
With some fluorophores such as 2 acetylanthracene or tryptophan a spectral shift to longer wavelengths (bathochromic shift or red shift) is observed in more polar solvents. As mentioned with AMC (please see below) the pH of a solution may also change the fluorescence properties of a fluorophore.
Fluorescence Quenching
Any process which decreases the fluorescence intensity of a given substance can be referred to as quenching. Several types of quenching processes can be distinguished. These include collisional and static quenching, as well as fluorescence resonance energy transfer (FRET). Collisional or dynamic quenching can be considered as a reduction in fluorescence intensity due to a collision of the quencher with the fluorophore in the excited state.
Upon contact the fluorophore returns to the ground state without light emission. One of the best known collisional quenchers which quenches almost all known fluorophores is molecular oxygen. It is therefore often required to remove dissolved oxygen to obtain reliable measurements.
In static quenching a non-fluorescent complex is formed between the quencher and the fluorophore. In contrast to both of these quenching processes, FRET does not require contact of the quencher with the fluorophore. The energy transfer occurs without the appearance of a photon.
Fluorescence Resonance Energy Transfer (FRET)
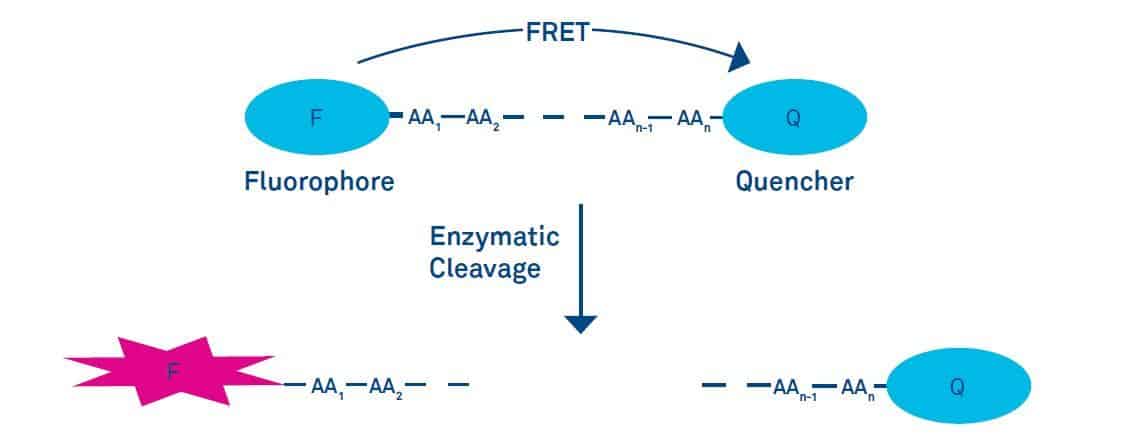
Fluorescence Resonance Energy Transfer (FRET)
Fluorescence energy transfer (FRET) is the transfer of the excited state energy of a donor to an acceptor without the emission of light. The energy transfer can be considered as an energy exchange of an oscillating dipole to a dipole with similar resonance frequency. In order to occur the emission spectrum of the donor has to overlap with the absorption spectrum of the acceptor, and the donor and acceptor have to be within a distance of 1-10 nm.
The energy transfer efficiency depends on the extent of the overlap of the emission spectrum of the donor with the absorption spectrum of the acceptor, the relative orientation of the donor and acceptor transition dipoles, and the distance r between donor and acceptor. The energy transfer efficiency decreases exponentially by r6. The distance at which the efficiency of energy transfer is reduced by 50% is a characteristic value for a given donor acceptor pair and is called the Förster distance R0.
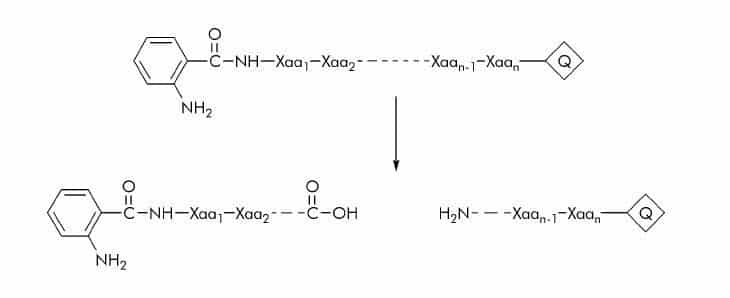
2-Aminobenzoyl or Anthraniloyl (Abz) Substrates
The Abz fluorophore is generally used in combination with a number of quenchers (Q) such as Dnp (2,4-dinitrophenyl), EDDnp (N-(2,4-dinitrophenyl)ethylenediamine), p-nitrophenylalanine, or 3-nitrotyrosine. Substrate cleavage can be detected at 420 nm using an excitation wavelength of 320 nm.
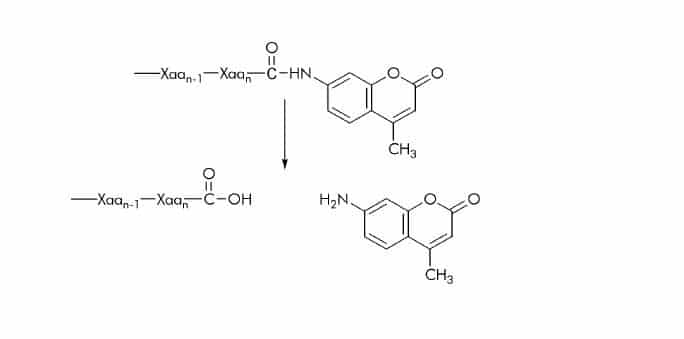
7-Amido-4-methylcoumarin (AMC) Substrates
Substrates with AMC attached to the C-terminal carboxyl group are commonly used for assaying carboxypeptidases. Cleavage of the amide bond between the peptide substrate and the AMC group results in the release of 7-amino-4 methylcoumarin which shows different excitation and emission spectra than the amide
bond linked fluorogen. It can be detected at 440-460 nm using an excitation wavelength of 360-380 nm. Like other aromatic amines AMC is partially protonated at low pH (< 5) but fully deprotonated at physiological pH. Therefore, at or near physiological pH its fluorescence spectrum is pH-independent.
5-[(2-Aminoethyl)amino]naphthalene-1-sulfonyl (EDANS) Substrates
In these substrates, the fluorescence of the EDANS group is generally quenched by the DABCYL (4-(4-dimethylaminophenylazo) benzoyl) group. The DABCYL group is usually conjugated to the Nterminus and the EDANS group (ε305nm = 24 700 M-1cm-1) attached to the C-terminus of the peptide substrate. Substrate cleavage can be detected at 490 nm using an excitation wavelength of 340 nm.
![5 [(2 Aminoethyl)amino]naphthalene 1 Sulfonyl (EDANS) Substrates](https://cdn.bachem.com/wp-content/uploads/2021/09/5-2-Aminoethylaminonaphthalene-1-sulfonyl-EDANS-Substrates.jpg)
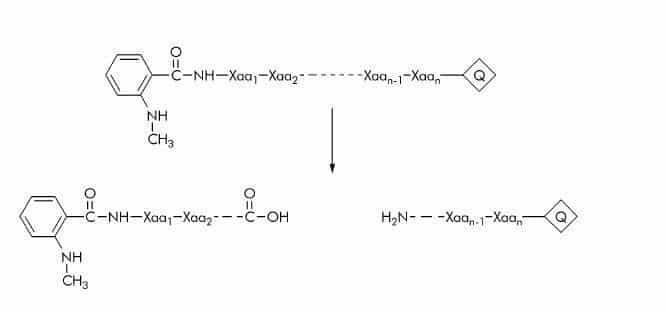
N-Methyl-anthraniloyl (N-Me-Abz) Substrates
N-Me-Abz substrates are generally used with Dnp as quencher (Q). The fluorescent group is either linked to the amino-terminal amino group or the ε-amino group of a lysine residue. Substrate cleavage can be detected at 440-450 nm using an excitation wavelength of 340-360 nm.
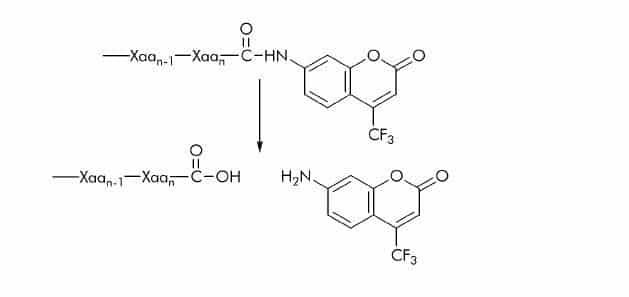
7-Amido-4-trifluoromethylcoumarin (AFC) Substrates
AFC substrates have the same basic structure as AMC substrates, the methyl group is replaced by a trifluoromethyl (CF3) group. The release of 7-amino-4-trifluoromethylcoumarin is usually detected at 495-505 nm using an excitation wavelength of 395-400 nm. AFC substrates exhibit similar sensitivity as AMC substrates.
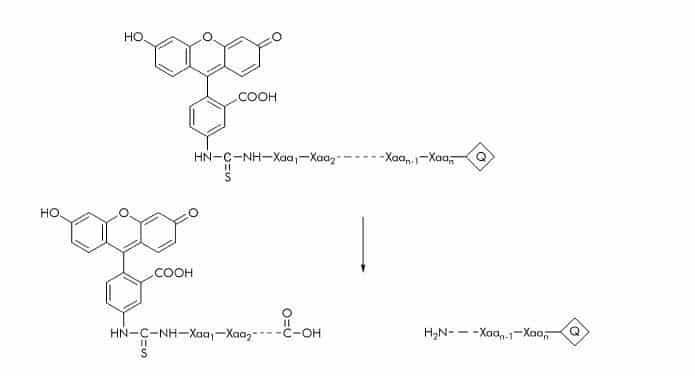
Fluorescein Isothicyanate (FITC) Substrates
Only few FITC substrates have been described. The FITC label can be quenched with Dnp. Substrate cleavage can be detected at 520 nm using an excitation wavelength of 490 nm.
Below you can find a list of common donor acceptor pairs used for the design of enzyme substrates.
| Donor | Acceptor (Quencher) | ||
|---|---|---|---|
| Abz[14] (2-Aminobenzoyl or Anthraniloyl) | Dnp (2,4-Dinitrophenyl) | ||
| Abz[32] (2-Aminobenzoyl or Anthraniloyl) | EDDnp (N-(2,4-Dinitrophenyl)ethylenediamine) | ||
| Abz [33] (2-Aminobenzoyl or Anthraniloyl) | p-Nitro-phenylalanine | ||
| Abz [34] (2-Aminobenzoyl or Anthraniloyl) | 3-Nitro-tyrosine | ||
| N-Me-Abz [17] (N-Methyl-anthraniloyl) | Dnp (2,4-Dinitrophenyl) | ||
| EDANS [23] (5-[(2-Aminoethyl)amino]-naphthalene-1-sulfonyl) | DABCYL (4-(4-Dimethylaminophenylazo)benzoyl) | ||
| FITC [35] (Fluorescein isothiocyanate) | Dnp (2,4-Dinitrophenyl) | ||
| Mca[29] ((7-Methoxycoumarin-4-yl)acetyl) | Dnp (2,4-Dinitrophenyl) | ||
| Trp [14] (Tryptophan) | Dnp (2,4-Dinitrophenyl) |
Table 3: Common Donor/Acceptor Pairs used at Bachem*
*the values listed are as reported in the cited literature.
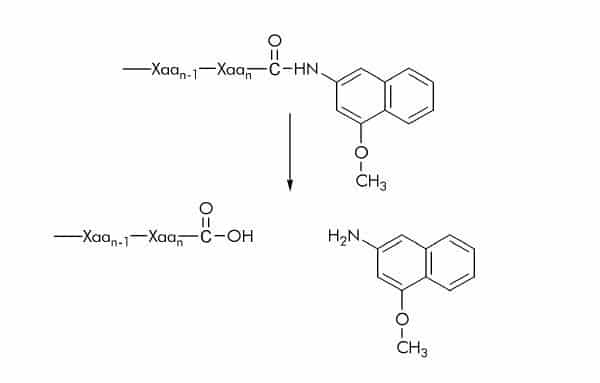
4-Methoxy-β-naphthylamide (4MβNA) Substrates
4MβNA substrates are commonly used for assaying protease activity. Cleavage of the amide bond between the peptide and 4MβNA results in the release of the fluorescent 4-methoxy-β- naphthylamine which can be detected at 410-440 nm using an excitation wavelength of 335-350 nm.
β-Naphthylamide (βNA) Substrates
As stated above (please see ‘Chromophores’) βNA substrates can either be used as chromogenic or fluorogenic substrates. The release of β-naphthylamine can be detected at 410-420 nm using an excitation wavelength of 320-340 nm.
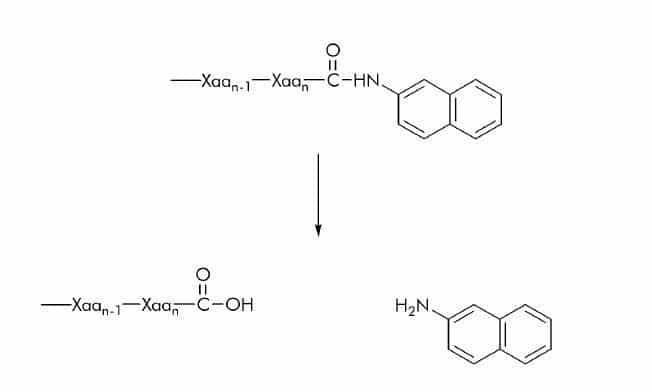
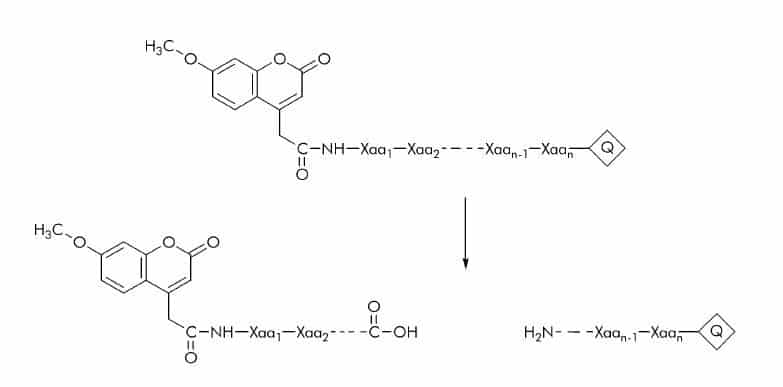
(7-Methoxycoumarin-4-yl)acetyl (Mca) Substrates
In this kind of substrates Mca is bound to an amino group (usually the N-terminal amino group) of a peptide sequence and quenched by Dnp (Q). The cleaved peptide fragment with the attached Mca group can be detected fluorometrically at 392 nm using an excitation wavelength of 325 nm.
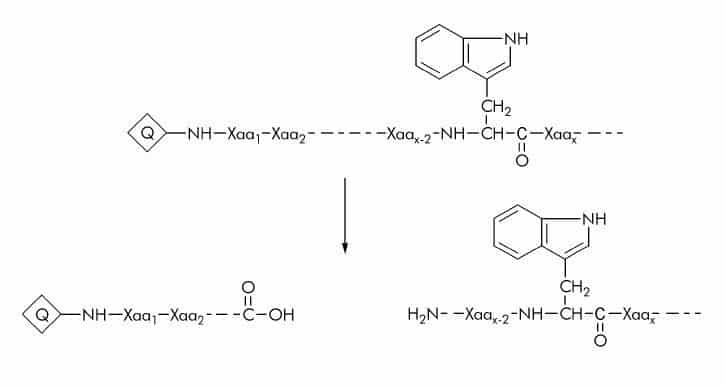
Tryptophan (Trp) Substrates
Tryptophan (like tyrosine and phenylalanine) is a fluorescent amino acid which has been used in a variety of substrates with Dnp as a quencher (Q). Substrate cleavage can be detected at 360 nm using an excitation wavelength of 280 nm.
Subscribe to our newsletter
"*" indicates required fields
References
[1] Y. Masui et al.
Synthetic substrates for vertebrate collagenase.
Biochem. Med. 17, 215-221 (1977)
(for Dnp-Pro-Gln-Gly-Ile-Ala-Gly-Gln-D-Arg OH (M-1340))
[2] A.R. Welch et al.
Purification of human matrilysin produced in Escherichia coli and characterization using a new optimized fluorogenic peptide substrate.
Arch. Biochem. Biophys. 324, 59-64 (1995)
(for Dnp-Arg-Pro-Leu Ala-Leu-Trp-Arg-Ser-OH (M-2205))
[3] Y.K. Li et al.
Mechanistic study of beta-xylosidase from Trichoderma koningii G-39.
J. Biochem. (Tokyo) 127, 315-320 (2000)
(for Dnp (pH 4.2))
[4] M. Abel et al.
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding site.
Biochem. J. 371, 997-1003 (2003)
(for Dnp (pH 7.2))
[5] H.E. Van Wart and D.R. Steinbrink
A continuous spectrophotometric assay for Clostridium histolyticum collagenase.
Anal. Biochem. 113, 356-365 (1981)
(for FA-Leu-Gly-Pro-Ala-NH2 (M-1385))
[6] M. Pank et al.
Hydrophobic interaction in thermolysin specificity.
FEBS Lett. 142, 297-300 (1982)
(for FA-Gly-Abu-NH2 (M-1835), FA-Gly-Val-NH2 (M-1820), and FA-Gly-Nva-NH2 (M-1830))
[7] M.N. Green et al.
The colorimetric determination of leucine aminopeptidase activity with L-leucyl-betanaphthylamide hydrochloride.
Arch. Biochem. Biophys. 57,458-474 (1955)
[8] P.M. Starkey and A.J. Barrett
Neutral proteinases of human spleen. Purification and criteria for homogeneity of elastase and cathepsin G.
Biochem. J. 155, 255-263 (1976)
[9] Y.-Q. Gu et al.
Overexpression, purification and biochemical characterization of the wound-induced leucine aminopeptidase of tomato.
Eur. J. Biochem. 263, 726-735 (1999)
[10] M. Zimmerman et al.
A new fluorogenic substrate for chymotrypsin.
Anal. Biochem. 70, 258-262 (1976)
[11] L. Visser and E.R. Blout
The use of p-nitrophenyl N-tert-butyloxycarbonyl-L-alaninate as substrate for elastase.
Biochim. Biophys. Acta 268, 257-260 (1972)
[12] C.R. Gentry-Weeks et al.
beta-Cystathionase from Bordetella avium. Role(s) of lysine 214 and cysteine residues in activity and cytotoxicity.
J. Biol. Chem. 270, 7695-7702 (1995)
[13] A.L. Niles et al.
Recombinant human mast cell tryptase beta: stable expression in Pichia pastoris and purification of fully active enzyme.
Biotechnol. Appl. Biochem. 28, 125-131 (1998)
[14] M.H. Cezari et al.
Cathepsin B carboxydipeptidase specificity analysis using internally quenched fluorescent peptides.
Biochem. J. 368, 365-369 (2002)
[15] L. Bourgeois et al.
Serpin-derived peptide substrates for investigating the substrate specificity of human tissue kallikreins hK1 and hK2.
J. Biol. Chem. 272, 29590-29595 (1997)
[16] K.N. Parameswaran et al.
Hydrolysis of gamma: epsilon isopeptides by cytosolic transglutaminases and by coagulation factor XIIIa.
J. Biol. Chem. 272, 10311-10317 (1997)
[17] D.M. Bickett et al.
A high throughput fluorogenic substrate for interstitial collagenase (MMP-1) and gelatinase (MMP-9).
Anal. Biochem. 212, 58-64 (1993)
[18] J.D. Cheng et al.
Promotion of tumor growth by murine fibroblast activation protein, a serine protease, in an animal model.
Cancer Res. 62, 4767-4772 (2002)
[19] M. Mogi and A. Togari
Activation of caspases is required for osteoblastic differentiation.
J. Biol. Chem. 278, 47477-47482 (2003)
[20] R. Bass et al.
Maspin inhibits cell migration in the absence of protease inhibitory activity.
J. Biol. Chem. 277, 46845-4688 (2002)
[21] B.Leiting et al.
Catalytic properties and inhibition of proline-specific dipeptidyl peptidases II, IV and VII.
Biochem. J. 371, 525-532 (2003)
[22] T.P. Dick et al.
Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants.
J. Biol. Chem. 273, 25637-25646 (1998)
[23] E.D. Matayoshi et al.
Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer.
Science 247, 954-958 (1990)
[24] A. Chersi et al.
Preparation and utilization of fluorescent synthetic peptides.
Biochim. Biophys. Acta 1034, 333-336 (1990)
[25] S.G. Rudyak and T.E. Shrader
Polypeptide stimulators of the Ms-Lon protease.
Protein Sci. 9, 1810-1817 (2000)
[26] J.M. Santana et al.
A Trypanosoma cruzi-secreted 80 kDa proteinase with specificity for human collagen types I and IV.
Biochem. J. 325, 129-137 (1997)
[27] S.L. Meyer et al.
Biologically active monomeric and heterodimeric recombinant human calpain I produced using the baculovirus expression system.
Biochem. J. 314, 511-519 (1996)
[28] T. Fukui et al.
A membrane-bound archaeal Lon protease displays ATP-independent proteolytic activity towards unfolded proteins and ATP-dependent activity for folded proteins.
J. Bacteriol. 184, 3689-3698 (2002)
[29] T. Kondo et al.
Activation of distinct caspase-like proteases by Fas and reaper in Drosophila cells.
Proc. Natl. Acad. Sci U S A. 94, 11951-11956 (1997).
[30] J. Lee et al.
Structure-activity study of LVV-hemorphin-7: angiotensin AT4 receptor ligand and inhibitor of insulin-regulated aminopeptidase.
J. Pharmacol. Exp Ther. 305, 205-211 (2003)
[31] A.L. Bulteau et al.
Proteasome inhibition in glyoxal-treated fibroblasts and resistance of glycated glucose-6- phosphate dehydrogenase to 20 S proteasome degradation in vitro.
J. Biol. Chem. 276, 45662-45668 (2001)
[32] D. Andrau et al.
BACE1- and BACE2-expressing human cells: characterization of beta-amyloid precursor protein-derived catabolites, design of a novel fluorimetric assay, and identification of new in vitro inhibitors.
J. Biol. Chem. 278, 25859-25866 (2003)
[33] M.V. Toth and G.R. Marshall
A simple, continuous fluorometric assay for HIV protease.
Int. J. Pept. Protein Res. 36, 544-550 (1990)
[34] K. Breddam and M. Meldal
Substrate preferences of glutamic-acid-specific endopeptidases assessed by synthetic peptide substrates based on intramolecular fluorescence quenching.
Eur. J. Biochem. 206, 103-107 (1992)
[35] H.J. Korting et al.
Fluorometric determination of the quality of FITC conjugates.
Virologie 28, 41-43 (1977)
Further References
J. Bergmeyer, M. Grassl, eds.
Methods of Enzymatic Analysis 3rd Edition, Vol. I, Fundamentals
Verlag Chemie GmbH, Weinheim (1983)
J. Bergmeyer, M. Grassl, eds.
Methods of Enzymatic Analysis 3rd Edition, Vol. II, Samples, Reagents, Assessment of Results
Verlag Chemie GmbH, Weinheim (1983)
J.R. Lakowicz
Principles of Fluorescence Spectroscopy
Plenum Press, New York (1983)
References
[1] Y. Masui et al.
Synthetic substrates for vertebrate collagenase.
Biochem. Med. 17, 215-221 (1977)
(for Dnp-Pro-Gln-Gly-Ile-Ala-Gly-Gln-D-Arg OH (M-1340))
[2] A.R. Welch et al.
Purification of human matrilysin produced in Escherichia coli and characterization using a new optimized fluorogenic peptide substrate.
Arch. Biochem. Biophys. 324, 59-64 (1995)
(for Dnp-Arg-Pro-Leu Ala-Leu-Trp-Arg-Ser-OH (M-2205))
[3] Y.K. Li et al.
Mechanistic study of beta-xylosidase from Trichoderma koningii G-39.
J. Biochem. (Tokyo) 127, 315-320 (2000)
(for Dnp (pH 4.2))
[4] M. Abel et al.
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding site.
Biochem. J. 371, 997-1003 (2003)
(for Dnp (pH 7.2))
[5] H.E. Van Wart and D.R. Steinbrink
A continuous spectrophotometric assay for Clostridium histolyticum collagenase.
Anal. Biochem. 113, 356-365 (1981)
(for FA-Leu-Gly-Pro-Ala-NH2 (M-1385))
[6] M. Pank et al.
Hydrophobic interaction in thermolysin specificity.
FEBS Lett. 142, 297-300 (1982)
(for FA-Gly-Abu-NH2 (M-1835), FA-Gly-Val-NH2 (M-1820), and FA-Gly-Nva-NH2 (M-1830))
[7] M.N. Green et al.
The colorimetric determination of leucine aminopeptidase activity with L-leucyl-betanaphthylamide hydrochloride.
Arch. Biochem. Biophys. 57,458-474 (1955)
[8] P.M. Starkey and A.J. Barrett
Neutral proteinases of human spleen. Purification and criteria for homogeneity of elastase and cathepsin G.
Biochem. J. 155, 255-263 (1976)
[9] Y.-Q. Gu et al.
Overexpression, purification and biochemical characterization of the wound-induced leucine aminopeptidase of tomato.
Eur. J. Biochem. 263, 726-735 (1999)
[10] M. Zimmerman et al.
A new fluorogenic substrate for chymotrypsin.
Anal. Biochem. 70, 258-262 (1976)
[11] L. Visser and E.R. Blout
The use of p-nitrophenyl N-tert-butyloxycarbonyl-L-alaninate as substrate for elastase.
Biochim. Biophys. Acta 268, 257-260 (1972)
[12] C.R. Gentry-Weeks et al.
beta-Cystathionase from Bordetella avium. Role(s) of lysine 214 and cysteine residues in activity and cytotoxicity.
J. Biol. Chem. 270, 7695-7702 (1995)
[13] A.L. Niles et al.
Recombinant human mast cell tryptase beta: stable expression in Pichia pastoris and purification of fully active enzyme.
Biotechnol. Appl. Biochem. 28, 125-131 (1998)
[14] M.H. Cezari et al.
Cathepsin B carboxydipeptidase specificity analysis using internally quenched fluorescent peptides.
Biochem. J. 368, 365-369 (2002)
[15] L. Bourgeois et al.
Serpin-derived peptide substrates for investigating the substrate specificity of human tissue kallikreins hK1 and hK2.
J. Biol. Chem. 272, 29590-29595 (1997)
[16] K.N. Parameswaran et al.
Hydrolysis of gamma: epsilon isopeptides by cytosolic transglutaminases and by coagulation factor XIIIa.
J. Biol. Chem. 272, 10311-10317 (1997)
[17] D.M. Bickett et al.
A high throughput fluorogenic substrate for interstitial collagenase (MMP-1) and gelatinase (MMP-9).
Anal. Biochem. 212, 58-64 (1993)
[18] J.D. Cheng et al.
Promotion of tumor growth by murine fibroblast activation protein, a serine protease, in an animal model.
Cancer Res. 62, 4767-4772 (2002)
[19] M. Mogi and A. Togari
Activation of caspases is required for osteoblastic differentiation.
J. Biol. Chem. 278, 47477-47482 (2003)
[20] R. Bass et al.
Maspin inhibits cell migration in the absence of protease inhibitory activity.
J. Biol. Chem. 277, 46845-4688 (2002)
[21] B.Leiting et al.
Catalytic properties and inhibition of proline-specific dipeptidyl peptidases II, IV and VII.
Biochem. J. 371, 525-532 (2003)
[22] T.P. Dick et al.
Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants.
J. Biol. Chem. 273, 25637-25646 (1998)
[23] E.D. Matayoshi et al.
Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer.
Science 247, 954-958 (1990)
[24] A. Chersi et al.
Preparation and utilization of fluorescent synthetic peptides.
Biochim. Biophys. Acta 1034, 333-336 (1990)
[25] S.G. Rudyak and T.E. Shrader
Polypeptide stimulators of the Ms-Lon protease.
Protein Sci. 9, 1810-1817 (2000)
[26] J.M. Santana et al.
A Trypanosoma cruzi-secreted 80 kDa proteinase with specificity for human collagen types I and IV.
Biochem. J. 325, 129-137 (1997)
[27] S.L. Meyer et al.
Biologically active monomeric and heterodimeric recombinant human calpain I produced using the baculovirus expression system.
Biochem. J. 314, 511-519 (1996)
[28] T. Fukui et al.
A membrane-bound archaeal Lon protease displays ATP-independent proteolytic activity towards unfolded proteins and ATP-dependent activity for folded proteins.
J. Bacteriol. 184, 3689-3698 (2002)
[29] T. Kondo et al.
Activation of distinct caspase-like proteases by Fas and reaper in Drosophila cells.
Proc. Natl. Acad. Sci U S A. 94, 11951-11956 (1997).
[30] J. Lee et al.
Structure-activity study of LVV-hemorphin-7: angiotensin AT4 receptor ligand and inhibitor of insulin-regulated aminopeptidase.
J. Pharmacol. Exp Ther. 305, 205-211 (2003)
[31] A.L. Bulteau et al.
Proteasome inhibition in glyoxal-treated fibroblasts and resistance of glycated glucose-6- phosphate dehydrogenase to 20 S proteasome degradation in vitro.
J. Biol. Chem. 276, 45662-45668 (2001)
[32] D. Andrau et al.
BACE1- and BACE2-expressing human cells: characterization of beta-amyloid precursor protein-derived catabolites, design of a novel fluorimetric assay, and identification of new in vitro inhibitors.
J. Biol. Chem. 278, 25859-25866 (2003)
[33] M.V. Toth and G.R. Marshall
A simple, continuous fluorometric assay for HIV protease.
Int. J. Pept. Protein Res. 36, 544-550 (1990)
[34] K. Breddam and M. Meldal
Substrate preferences of glutamic-acid-specific endopeptidases assessed by synthetic peptide substrates based on intramolecular fluorescence quenching.
Eur. J. Biochem. 206, 103-107 (1992)
[35] H.J. Korting et al.
Fluorometric determination of the quality of FITC conjugates.
Virologie 28, 41-43 (1977)
Further References
J. Bergmeyer, M. Grassl, eds.
Methods of Enzymatic Analysis 3rd Edition, Vol. I, Fundamentals
Verlag Chemie GmbH, Weinheim (1983)
J. Bergmeyer, M. Grassl, eds.
Methods of Enzymatic Analysis 3rd Edition, Vol. II, Samples, Reagents, Assessment of Results
Verlag Chemie GmbH, Weinheim (1983)
J.R. Lakowicz
Principles of Fluorescence Spectroscopy
Plenum Press, New York (1983)